Bricklet Announcement
Mac Cowell and I have been working on a new project, coined Bricklet. Bricklet is an open and extensible platform for storing and sharing standardized synthetic biology parts with the goal of fostering a rich ecosystem of synthetic biology software.
Bricklet currently consists of:
- A proposal for a Part Description Language
- A proposal for a Parts Sharing Framework that supports a web of registries, selective publication, document revisioning, and provenance/attribution.
Our intent is to implement ideas from the synthetic biology community and the BioBricks Technical Standards Working Group. We want to exercise these ideas with the hope of gaining insight into both their advantages and limits, with the intent to iterate in the future. Eventually, we may like to submit our ideas as patches to a project like Brickit to reuse existing functionality and build development mindshare.
We are tracking the requirements, design, and implementation on the Bricklet page at Google Code page. Mac and I will be presenting our progress at the Standards and Specifications in Synthetic Biology Workshop at the end of this month.
BarCampRochester3
Last weekend, I has the pleasure to attend BarCampRochester which was a great time. Thanks to the organizers and sponsors for making this happen!
I gave a session that introduced synthetic biology, iGEM, their motivations, and surrounding issues. I’ve uploaded my slides in PDF (1.5MB): Synthetic Biology at BarCampRochester.
Here are a few takeaways from some of the sessions I attended:
There was a great session and discussion about intellectual property: copyright, patents, and trademarks – in particular how these apply to software and why patents aren’t necessarily evil (although the patent duration is surely out of touch with the speed of the software market). I’ll be reading more about Open Innovation, with an eye toward its applicability to both software and science.
Al Biles led a brainstorming session about the nature of creativity and what it means to be creative. This was an open-ended discussion with an exploratory nature, and was quite enjoyable. Al differentiated between P-creativity, which is an act that is original from an indivudal’s perspective, and H-creativity, which is an act that is original with respect to all known history. He also recommended Margaret Boden’s “Dimensions of Creativity.”
I learned about the difficultly of accessing supposedly open governmental data in the US due to its distribution in proprietary or obtuse formats. Consider a database that is made accessible by taking screenshots from within the Oracle admin tool, printing these out, scanning them back in, and distributing the lot as a PDF. Fighting the good fight, there are projects like those at the Sunlight Foundation that focus on making this data more readily accessible.
Then, there are projects like EveryBlock, which collates such data and lets you filter it by location, so you can learn about happenings in your neighborhood from crimes to business licensing to permit issuances. This is a great trend, and I hope to see it grow both in the domain of making data accessible and making it useful.
Following up on the political theme, I was in a thought-provoking session called “So you want to become a lobbyist?” that took a look at the importance of some of the “nuts and bolts” political issues like redistricting, and how effective grassroots movements are on a local scale (the consensus: very effective). Remy made an interesting point that grassroots means person-to-person, whether that’s door-to-door or online.
Sam & Katie gave a refreshing talk about relationship branding: 2 cool kids = 1 cool brand: thinkskinc.com.
Justin Thorp has a great post that he wrote post-BarCamp about starting your personal branding in college that is spot on. Having attended a fair number of conferences, I was also caught slightly off-guard by the lack of biz card trading. Go go day job plug for business cards!
Finally, if you’re in the Rochester, NY area, definitely check out the Society of Lectors, a group of folks who hold regular meetings to give BarCamp style presentations on a wide gamut of topics. Go brush up on your presentation skillz, and learn something new!
Managing Your Research
There are a few activities that, arguably, comprise the bulk of science.

They are, of course, not linear.

And each one generates many artifacts.

…many, many, many artifacts.

Wouldn’t it be nice to keep track of all these? (Especially in a distributed team!)
Traffic Is a Routing Problem
2 ideas and then 1 idea:
1. GPS devices need more information about real-time traffic information. The best source for this is other GPSes that are also stuck in traffic. If they could communicate, they could intelligently route traffic to optimally spread traffic over primary and alternate routes. There are products that do this. iPhone and Android would also be excellent candidates.
2. To share this information, they need to be networked. They can form an ad-hoc wifi network when near one another and nodes with internet connections like internet phones and nodes near municipal wifi (which could be embedded into traffic lights?) can act as up/down links to a central database of traffic information.
3. In lieu of available real-time information, devices could sync with the central database by docking. A Bluetooth-enabled GPS could send its daily traffic recordings to your phone, which syncs itself to your computer when you carry it inside to home or work, which communicates this information to the internet. Pattern recognition is applied, and then traffic patterns are downloaded via the same channels to your GPS. It can use the traffic patterns to avoid areas of likely congestion during your route. (And you could cut out the middleman if your phone is your GPS.)
Lolscience
I actually had to register for a LiveJournal account today (ack), but it was for a good cause: to post to lolscience.
Cell Signaling, Computer Architecture, Functional Programming, and AI Blackboards
While thinking about synthetic biology, I find it useful to identify analogues of biological systems in a discipline I am already familiar with, computer science. This helps me better understand the new concept and can also raise questions about the new system that already have discussions around them in the CS world. Of course, the fidelity of such metaphors is not 100%, so I have to take care to ground such discourses with actual biology, but it’s been great for brainstorming so far.
Here are a few such metaphors.
Computer Architecture
One challenge in building composable biological systems for effective abstraction hierarchies is cell signaling crosstalk. One issue at play is a possible shortage of signal carriers; if you need ten different processes to occur at the same time and to act orthogonally, you will need ten signal carriers. Much as CPUs have a finite number of registers and data buses have finite width, there are only so many standardized signal carriers that standardized parts currently accept. Certainly, more carriers could be designed, but there may be ways around this. In computer architecture, the register shortage can be addressed by temporarily saving register data into memory, and the bus issue can be addressed with a stateful multiplexing approach. Perhaps stateful biological systems such as the repressilator or a push-on push-off switch have something to offer here.
Functional Programming
The other, more subtle, crosstalk issue is that of unintended side effects. When considering engineered biological devices, there may be side effects outside of the well-characterized and intended inputs and outputs, so the abstractions leak – figuratively and literally. These issues are also present in highly parallel computing environments when a shared resource such as a location in memory is operated upon by many processes. One must take care to ensure that the processes cooperate to ensure that they do not tread upon one another. There are many paradigms for approaching concurrent software, and it is becoming increasingly apparent that a system of threads, locks, and mutexes quickly gets difficult, if not impossible, to keep track of. Interesting discussions of other approaches:
- Software Transactional Memory (video)
- Beautiful Concurrency (PDF) is also about STM
- Actors that Unify Threads and Events (PDF)
- Simon Peyton Jones’ talk on nested data parallelism
- The Next Mainstream Programming Language: A Game Developer’s Perspective (PPT), particularly pages 49-56
A key element in many of these approaches to concurrency is a lack of shared state and, by association, lack of side effects. Since intracellular signaling systems are inherently parallel, two operations may only be reliably executed in tandem if they have minimal-to-no side effects (or, more realistically, minimal, orthogonal, well-characterized effects). My point here is not to imply that there is a concurrent programming paradigm that can be transferred to synthetic biology to solve the side effect issue. I mean to illustrate that, as we engineer larger systems, it will be crucial to minimize unknown side effects of synthetic biology constructs with high-quality characterization.
I realize that I am first suggesting stateful systems, immediately followed by a call for referential transparency (the property of being without formal side effects), often conflated with statelessness. It’s important to remember that, taken as an abstract concept, referentially transparent (side-effect free) operations can maintain internal state, so long as that state does not leak out of the operation, be it a computation or intracellular signalling pathway.
Also, something on my TODO list is to check out cell-free systems, and consider their applicability to the crosstalk issue:
- Imperial College’s iGEM 2007 work on Cell-Free Systems
- Construction of an in vitro bistable circuit from synthetic transcriptional switches
AI Blackboards
Lastly, it also occurred to me that intracellular signaling systems are similar to the artificial intelligence blackboard architecture and, as such, it could be useful as a thought model. I’ve not pursues this idea very much, although it appears that some folks have made the same connection in the context of modeling: Modelling intracellular signalling networks using behaviour-based systems and the blackboard architecture.
Bio Eye for the CS Guy (or Gal)
Lately, as a result of my fascination with Synthetic Biology, I have been reading biology and bioinformatics references voraciously. More on Synthetic Biology to come, but I encourage you to read up on it. It suffices to say that it greatly appeals to me, coming from a Computer Science and engineering background. When you throw “refactoring” and “bacteriophage” into the same paper title, or mention languages and grammars for programming DNA, you’ve got my attention.
To my surprise, I have found several resources that introduce biological concepts to readers with just such a background. In the interest of sharing this information over primping and editing this in Mephisto admin, here’s a work-in-progress of said list.
I have partially or fully read, and recommend:
- Cohen, Jacques. Bioinformatics: An Introduction for Computer Scientists . (PDF)
- Cohen, Jacques. Computer Science and Bioinformatics. (PDF)
- Cohen, William. A Computer Scientist’s Guide to Cell Biology. (book, publisher’s site)
- Hunter, Lawrence (editor). Artificial Intelligence and Molecular Biology.. (full text in HTML)
For the last entry, especially note chapters 1, “Molecular Biology for Computer Scientists,” and 2, “The Computational Linguistics of Biological Sequences .”
This last one is not related to programming, but is an amazing introduction to cellular biology that I would highly recommend for its fantastic illustrations and readable prose, whether you are familiar with the material or not:
- Goodsell, David. The Machinery of Life (book, Amazon referral-less link)
Sardines
Sardines is an experiment in organizing a tiered, distributed wiki that is motivated by Open Science.
Imagine research that takes place in a lab and is recorded and documented on an electronic platform such as a blog or wiki. It’s reasonable to conceive that the researcher may want to keep a closer hold on their findings for a short time in order to polish and confirm, before releasing it into the open.
This being said, it would be nice if the data, before being released publicly, were available within the entire research lab or institution. It’d also be great if the publication process seemed relatively seamless and that the interfaces for local private edits and global public edits be similar. To cap it all off, people “downstream” (i.e. in the lab) should automatically get updates when changes are made available “upstream.”
Example
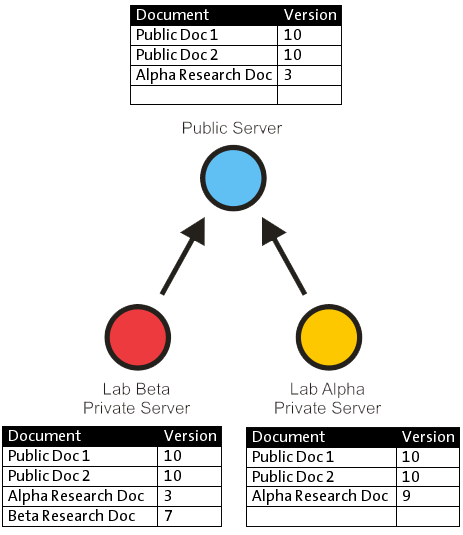
Distributed document versioning
In this example, there is one public server and two private servers; labs Alpha and Beta each have a private instance of the server running so that they are assured of privacy. Lab Alpha has published version 3 of its research, although they have an internal copy which is more recently updated. The published version is available on the public server and is made known to Lab Beta’s server. Lab Beta has private research which it has not yet released.
Notes
It may very well be the case that this idea constitutes “too much software,” and a simple published-state property on a central wiki would suffice.
Ruby Type Inference and Code Completion for RDT
I gave a talk at RubyConf 2006, detailing my project for the Google Summer of Code 2006. I worked on type inference for the purpose of code completion in the Ruby Development Tools Eclipse plugin with Chris Williams as my mentor. Chris has gone on to work with Aptana on RadRails.
You can read the archives of my project blog.
